Development of a Method for Quick Analysis of Conformational Changes in Proteins — Expectations for the Development of New Drugs —
Research Press Release | December 14, 2015
| Press Release | ||
|---|---|---|
| Key Points |
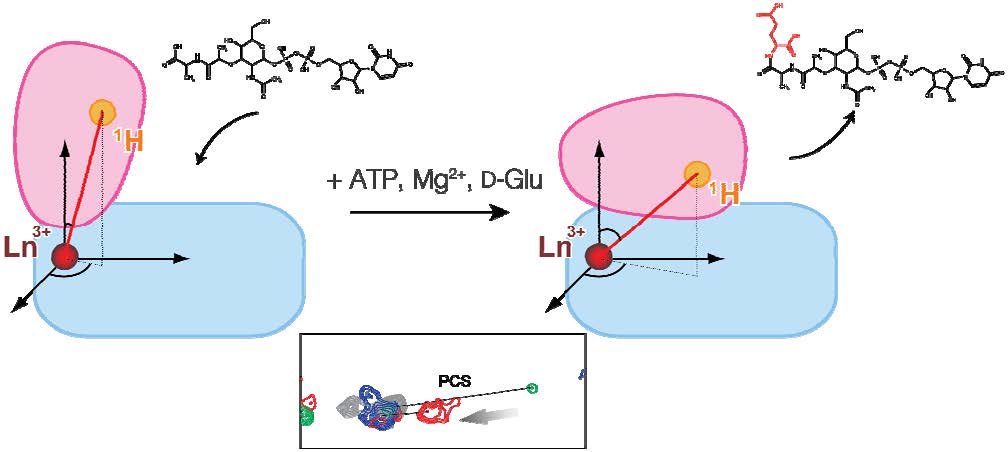
・We have monitored the conformation of MurD, an enzyme related to the synthesis of the bacterial cell wall, and succeeded in observing its conformational changes in detail. ・MurD has received attention as a new stepping-stone to antibacterial agents. ・This success is expected to lead to the development of new drugs. |
|
|
Outline |
Proteins, which perform many important functions in the body, fulfill their functions by changing their shapes (conformations) flexibly. It is known that MurD, an enzyme related to the synthesis of the bacterial cell wall, undergoes significant conformational changes to fulfill its function, but the details have not been understood. We have succeeded in monitoring the conformation of MurD by using the technique of nuclear magnetic resonance (NMR), and in making detailed observations of how the binding to a low-molecular substrate triggers the conformational changes of MurD during the enzymatic reaction. MurD has received attention as a new stepping-stone to antibacterial agents, so the results obtained by this research are expected to lead to the development of new drugs. This research was performed under a grant-in-aid from the Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program of the Ministry of Education, Culture, Sports, Science and Technology. |
|
|
Terms |
1 MurD: A enzyme involved in the synthesis of the peptidoglycan layer that forms the bacterial cell wall. It is an enzyme unique to bacteria, so it has received attention as a stepping-stone to anti-bacterial agents. 2 Paramagnetic lanthanide ion: Among the elements of the lanthanide series that constitute the rare-earth elements, those that show paramagnetism in their ionic forms. By observing paramagnetic effects from the NMR signals of proteins, one can capture long-range structural information of the protein from the lanthanide ion. 3 Nuclear magnetic resonance (NMR): A method to observe nuclear spins in a protein in a strong magnetic field, to investigate structure, dynamics, and interaction of proteins, at atomic resolution. |
|
| Inquiries |
Coordinating Office, Future Drug Discovery and Medical Care Innovation Project, Hokkaido University Publicity liaison: Ms. Wada Phone: +81-11-706-7798 FAX: +81-11-706-7799 E-mail: innovation@cris.hokudai.ac.jp |
|
|
Japanese Link |
タンパク質の立体構造変化を迅速に解析する手法を開発 ~新規薬剤開発への展開へ期待~ (11.20.2015) | |
| Publications | Ligand-driven conformational changes of MurD visualized by paramagnetic NMR, Scientific Reports (11.19.2015) | |
