DNA computing brings molecular robot a step closer to reality
Research Highlight | April 26, 2019
This article is an excerpt from the research magazine “Tackling Global Issues vol.1 Soft Matter: Material of the Future.” Click here to see the table of contents.
In 2016, Akira Kakugo and his research team introduced a novel approach for sensing surface mechanical deformation of a soft material. One year later, Kakugo had not only the sensor but also a processor and an actuator – the three essential components for producing a molecular robot.
To measure the surface deformation of a soft material, the team utilized cellular proteins called microtubules and kinesins. Microtubules are lament-shaped proteins that serve as railways in the cellular transportation system, while kinesins are motor proteins that run on these railways. The team, however, took a reverse approach: kinesins are fixed on the surface of a soft material to allow microtubules to move around on them, like railways running on motors. This method allowed the team to successfully measure the surface deformation by detecting changes in the moving patterns of microtubules. “They move randomly but show different patterns as the surface deforms. The patterns reflect the direction and degree of deformation,” Kakugo explained.
“A moving sensor was a novel concept when we published the paper in 2016,” Kakugo said. “Since we got molecules that move autonomously and randomly in a test tube, the next question became how to control their random movement to produce collective motion.”
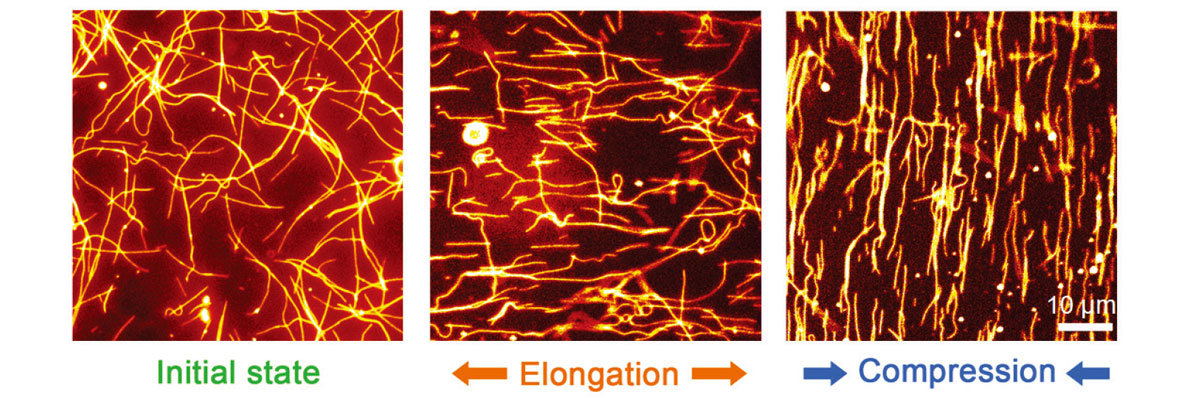
The team successfully measured the surface deformation by observing changes in the patterns of moving microtubules. (Inoue D. et al., Nature Communications, Oct 3, 2016)
Video: Fluorescent-microtubules run randomly on the surface of a soft material (Inoue D. et al., Nature Communications, October 3, 2016)
Let DNA take control
“Collective motion” is the key concept when they guide moving molecules. “When molecules act as a group, new properties that cannot be possessed by individual molecules appear, which we call ‘emergence’,” Kakugo explained. Such new properties are exemplified by division of labor, enhanced robustness and effectiveness, and flexibility that allows them to swiftly respond to a changing environment. Importantly, collective motion will enable molecules to work as an actuator, a device for moving something, when built in a molecular robot.
After testing several options, the team devised the idea of using a DNA computer to control the motion of microtubules. “Combining DNA with microtubules sparked a breakthrough in our research,” Kakugo said. A molecular machine built on such a concept had been nonexistent when they reported the discovery in early 2018.
But how can DNA, an essential element of life, control the physical movement of other molecules? In the team’s experiments, DNA – homophilic molecules that bind to each other when they have complementary sequences – are attached simply to microtubules. Also added were DNA molecules, or input DNA, that intermediate interactions between DNA-attached microtubules.
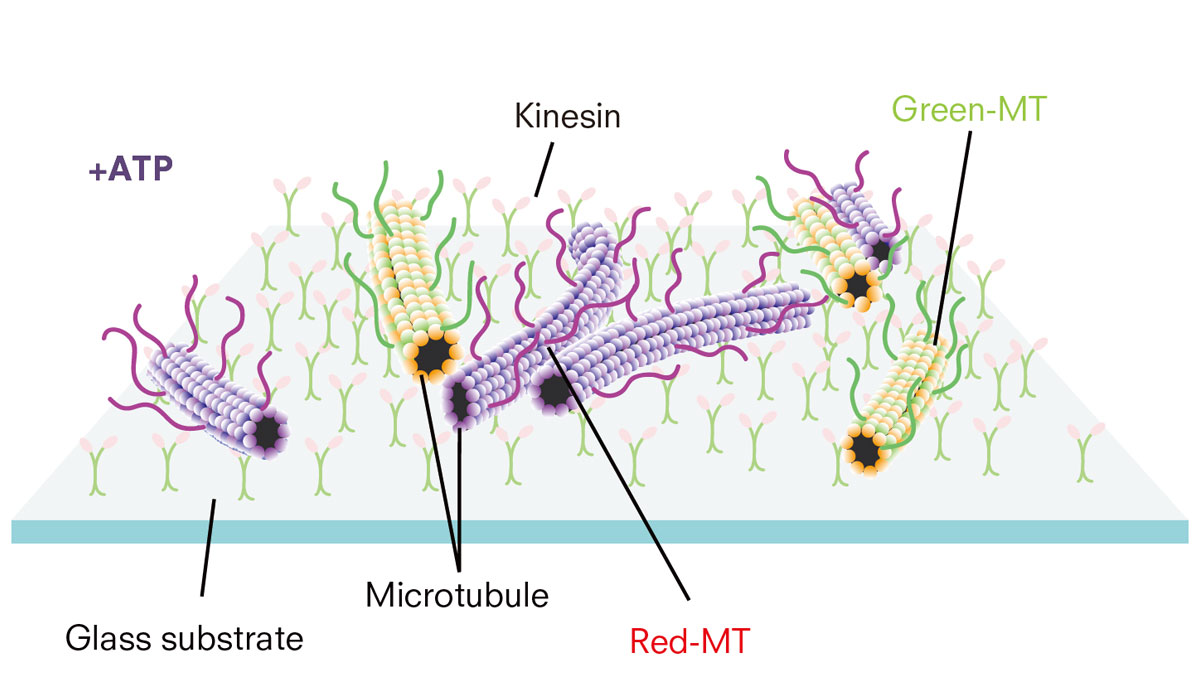
Microtubules (MT) interact with each other through DNA molecules while moving around on the kinesin motors. Microtubules were labelled with green and red fluorescence for observation. (Keya J. J. et al., Nature Communications, January 31, 2018)
When this system was introduced, DNA-attached microtubules stopped moving randomly but started swarming and self-organizing into different shapes that exhibited various patterns of movement. “We could make different patterns of collective motions by changing the programs in the DNA sequence, therefore changing the interactions between them,” explained Kakugo. They also showed the swarming motion could be modulated by changing the rigidity of microtubules.
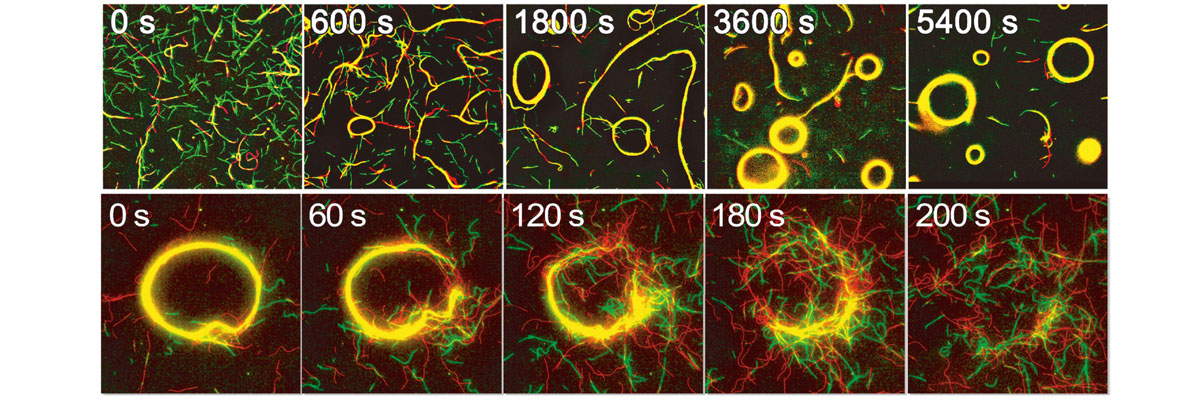
The exible (low rigidity) microtubules formed swarms with circular motions (top). A swarm of microtubules was disassembled by adding input DNA with a certain sequence (bottom). (Keya J. J. et al., Nature Communications, January 31, 2018)
Video: Swarming behavior of microtubules programmed by DNA computing (Keya J. J. et al., Nature Communications, January 31, 2018)
Focusing on collective motion of birds

The team applied the Boids model to DNA computing, an artificial life simulation to replicate the aggregation motion of a flock of birds, herd of land animals or school of fish.
The breakthrough was made possible mainly because of his application of the Boids model, an artificial life simulation to replicate the aggregation motion of a flock of birds, herd of land animals or school of fish. Kakugo introduced the concept to DNA computing for the first time.
“DNA-attached molecules acknowledge their comrades through contact and the exchange of information,” Kakugo said, showing a video to demonstrate how a flock of molecules make translational motions or rotational motions – clockwise or anticlockwise – like fish. To realize the flocking behavior, three factors in the Boids model – attraction, repulsion and alignment – had to be programed in the DNA sequence. His team also added azobenzene to the system, a chemical that binds to DNA and changes its structure in response to light. This enabled them to switch the DNA computer on and off by using light.
Learning from a flea
Kakugo is now trying to build a system in which actuators are hierarchically ordered to enhance their scalability, complexity of movements and intelligent behavior. “The concept might be easier to understand if you imagine the Transformers or Microbots (in Big Hero 6) featured in U.S. movies,” he said.
To attain this goal, he has to increase the number of sensors from the current one, a light sensor, to make it behave intelligently. A temperature sensor is a prime candidate for the next step.
“A flea is a primitive creature, but it has three sensors, for light, temperature and butyric acid,” Kakugo said. “It climbs up a tall tree when sensing light, jumps off the tree when sensing butyric acid in animal perspiration, and bites when sensing a certain temperature on the animal. That’s how fleas can suck blood. With only three sensors, they appear to be behaving intelligently.”
More challenges lie ahead
Although research into molecular motors has made considerable progress, Kakugo has identified several challenges that must be overcome before they are put into practical use.
First, researchers must boost the robustness of biological materials for use in medical fields and industries. “Present actuators based on biomolecules are prone to degrade, so we might be able to start with a disposable motor for practical use to spearhead ensuing research,” Kakugo said.
It is not easy, however, to engineer matter that moves actively in the current academic framework. The discipline of physical chemistry handles matter with equilibrium dynamics, but Kakugo handles active matter that moves autonomously with nonequilibrium dynamics, in which energy and substances come and go. “I think we must establish a new interdisciplinary framework for active matter if we want to see major advances in this field,” he said. “To realize life-like behavior of molecular robots, incorporating algorithms of artificial intelligence would be particularly exciting.”
Click here to see the table of contents.


