Novel method developed to extract molecular states and their network from noisy single molecule time series data
Research Press Release | March 23, 2015
| Press Release | ||
|---|---|---|
| Overview |
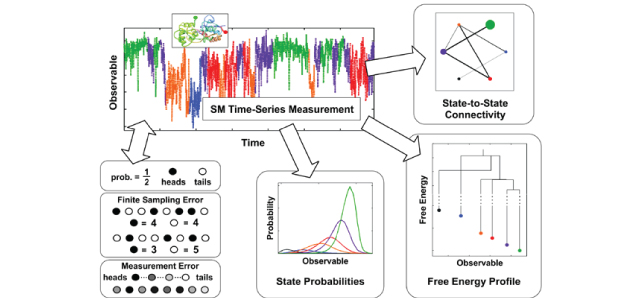
Methods from statistics and information theory were used to quantify errors in the time-series measurements and extract valuable and detailed information about biological systems. It was applied to experimental single-molecule (SM) time-series acquired for a neurological protein known as the AMPA receptor, which mediates the transmission of calcium ions through the central nervous system. AMPA is the most abundant protein in our nervous system and has been implicated in disorders such as depression and epilepsy. Uncovering vital information about the AMPA receptor has deepened understanding of how the protein moves in response to various biological stimuli, which is also expected to be applicable for medical applications. Read on here: |
|
| Inquiries |
Tamiki Komatsuzaki, Professor, Laboratory of Molecule & Life Nonlinear Sciences, Research Institute for Electronic Science, Hokkaido University TEL & FAX: +81-11-706-9434 E-mail: ries-sec@es.hokudai.ac.jp |
|
|
Japanese Link |
ノイズを含むデータから分子の状態とそれらのネットワークを 抽出する新しい手法の開発にはじめて成功 | |
| Publications | Error-based Extraction of States and Energy Landscapes from Experimental Single-Molecule Time-Series, Scientific Reports (3.17.2015) | |
