Revealed: How SARS-CoV-2 evades our immune system
Research Press Release | December 07, 2021
Joint release by Hokkaido University and Texas A&M University
Scientists at Hokkaido University and Texas A&M University have identified a key mechanism used by the SARS-CoV-2 virus to evade host immune systems.
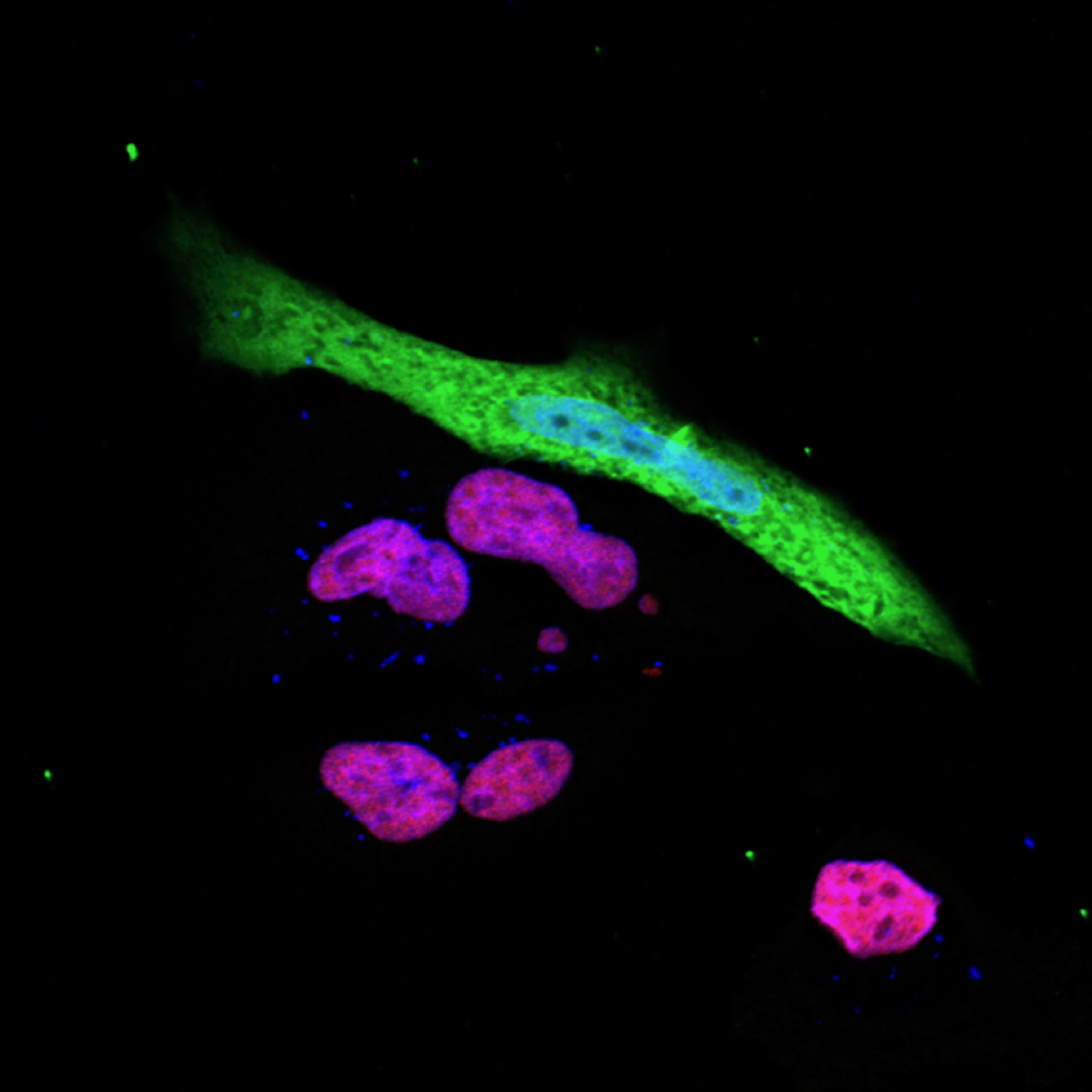
The expression of the immune response gene (red purple) is suppressed in SARS-CoV-2 (green) infected cells (Photo: Ji-Seung Yoo, Koichi Kobayashi).
Researchers in Japan and the United States have found SARS-CoV-2 can knock out an important molecular pathway linked to an immune complex called MHC class I. The finding should help scientists better understand how COVID-19 infection takes hold.
“Our discovery reveals how the virus can evade the human immune defense system and might help to explain why the pandemic has been so severe,” says Hokkaido University and Texas A&M University immunologist Koichi Kobayashi, who led the study. “The mechanisms we identify may provide new molecular targets for drug discovery.”
The scientists used a bioinformatics approach to look at how SARS-CoV-2, the virus that causes COVID-19, changes gene expression in the immune systems of COVID-19 patients compared to uninfected individuals. This is a useful way to look into the function of complicated cell signalling pathways that trigger immune responses to fight off harmful bacteria and viruses.
MHC (major histocompatibility complex) class I molecules are a central weapon in the immune response against viruses. When a virus infects a cell, the cell facilitates the expression of viral antigens on the surface of infected cells, drawing the attention of immune cells called cytotoxic T cells. These immune cells zero in on and destroy the infected cells, together with the invading virus inside them.
In addition to analysing gene expression in COVID-19 patients, the research team also infected human cell lines with the SARS-CoV-2 virus to validate their findings.
The results showed that a protein from the SARS-CoV-2 virus, called ORF 6, suppresses a host cell protein, called NLRC5, responsible for activating the MHC class I pathway.
The study showed this happens in two ways. ORF6 hampers cell signalling, which turns off the expression of NLRC5. ORF6 also blocks the function of NLRC5.
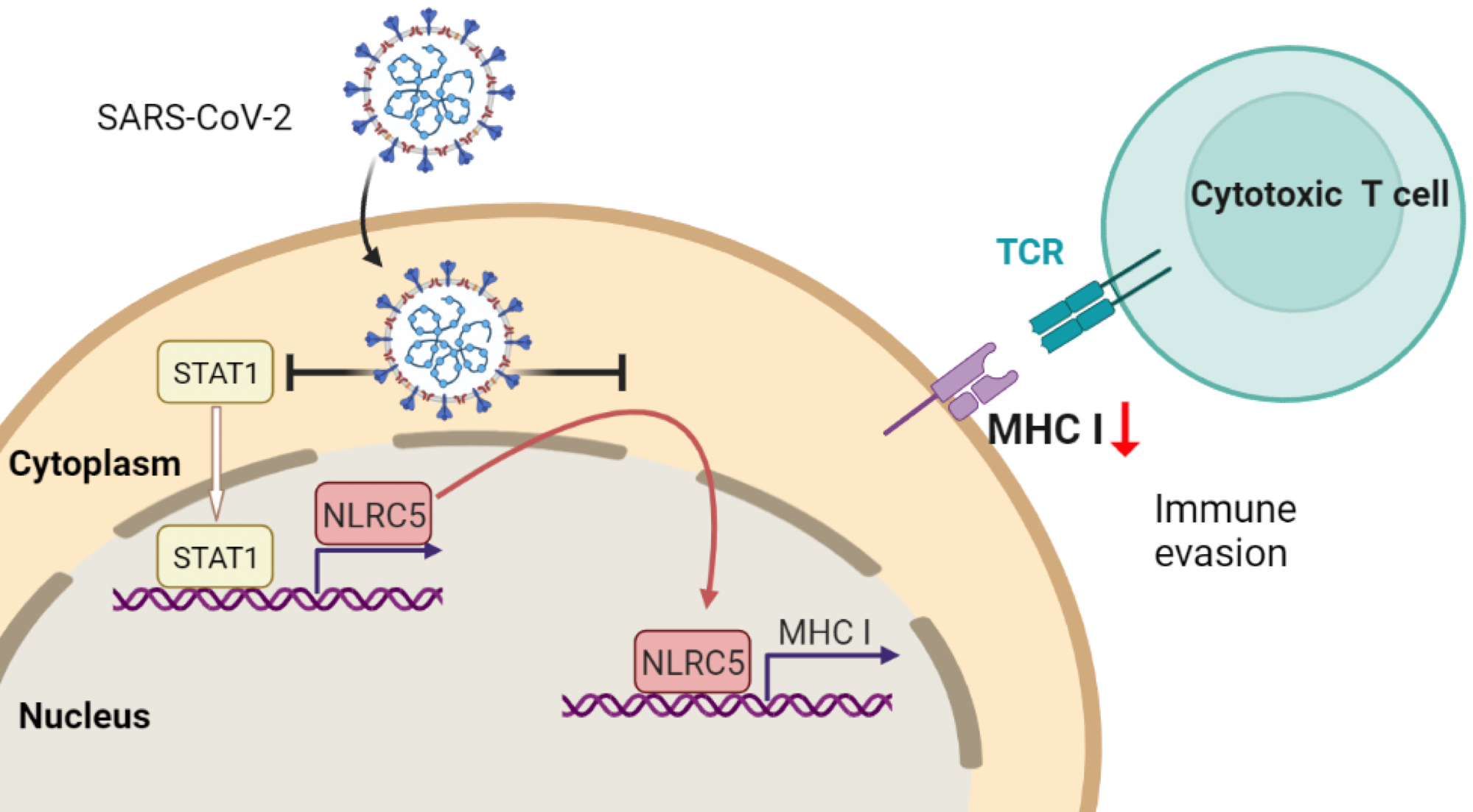
SARS-CoV-2 escapes from immune responses by cytotoxic T cells via impaired MHC-I expression which is caused by reducing both the amount and function of NLRC5 (Baohui Zhu, Koichi Kobayashi).
Other infectious viruses, including HIV and MERS, are known to also target the MHC class I pathway. Researchers believed that SARS-CoV-2 probably did as well, but this study is the first to unravel the mechanism.
“Without the activation of the MHC class I pathway, viruses in the infected cells are essentially hidden from the immune system. That helps to explain why SARS-CoV-2 virus persists in the body and why it keeps infecting others, leading to the pandemic,” Kobayashi says.
Further research could help find and test drugs that block the activity of the ORF6 viral protein, to restore host cell ability to activate the major histocompatibility complex. If successful, such drugs could encourage the host immune system to clear the virus itself, effectively boosting immune responses.
The press release from Texas A&M can be found here.

Steven X. Cho, Ji-Seung Yoo, Koichi Kobayashi, Ryota Ouda (front row, from the left), and Baohui Zhu (middle row, center left), contributing authors from the Department of Immunology, Graduate School of Medicine, Hokkaido University (Photo: Koichi Kobayashi).

(From the left) Yasuko Orba, Hirofumi Sawa, Michihito Sasaki, contributing authors from the Division of Molecular Pathobiology, International Institute for Zoonosis Control, Hokkaido University (Photo: Hirofumi Sawa).

Yusuke Kasuga (Graduate School of Medicine, Hokkaido University), a contributing author of the paper (Photo: Yusuke Kasuga).
Original Article:
Ji-Seung Yoo, et al. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nature Communications. November 15, 2021.
DOI: 10.1038/s41467-021-26910-8
Funding:
This work was supported by grants from the American Lung Association (LCD-507710), National Multiple Sclerosis Society, CSTR, TAM Genomics, Japan Society for the Promotion of Science (JSPS; 18H06135, 20K21511, 19K23837, 21K15445, 19K16681), SENSHIN Medical Research Foundation, Bristol Myers Squibb, Takeda Science Foundation, Kaketsu-ken foundation, Kobayashi Foundation, the Japan Program for Infectious Diseases Research and Infrastructure (JP20wm022500) from Japan Agency for Medical Research and Development (AMED), and The Hitachi Global Foundation.
Contacts:
Professor Koichi S. Kobayashi
Department of Immunology
Graduate School of Medicine
Hokkaido University
Tel: +81-11-706-5056
Email: kskobayashi[at]med.hokudai.ac.jp
Department of Microbial Pathogenesis and Immunology
Texas A&M Health Science Center
Email: kobayashi[at]tamu.edu
Professor Hirofumi Sawa
Division of Molecular Pathobiology,
International Institute for Zoonosis Control
Hokkaido University
Tel: +81-11-706-5185
Email: h-sawa[at]czc.hokudai.ac.jp
Professor Paul de Figueiredo
Department of Veterinary Pathobiology
Texas A&M University
Tel: +1-979-436-0699
Email: pjdefigueiredo[at]tamu.edu
Sohail Keegan Pinto (International Public Relations Specialist)
Public Relations Division
Hokkaido University
Tel: +81-11-706-2185
Skype: hokudai.pr1
Email: en-press[at]general.hokudai.ac.jp
Christina Sumners
Texas A&M University
Tel: +1-979-436-0616
Email: christina.sumners[at]tamu.edu
