Spotlight on Research: Uprooting the Tree of Life
Research Highlight | July 25, 2018
“I am not a mathematician,” says Professor Kei Kitahara.
The Hokkaido University molecular microbiologist repeats this phrase frequently when discussing his career as an academic. It is this fact that peaked his interest in microbiology in the first place; as a high-school student, a teacher asked his class how long it would take a single bacterium to become 1,000 bacteria, assuming they duplicate every 20 minutes. “I mistakenly answered 20,000 minutes, thinking simply to multiply 20 by 1,000” he explains. When informed the real answer was closer to 200 minutes, Kitahara remembers being amazed that life was capable of replicating itself so quickly, and from this moment he decided to embark on a career in microbiology.
It was also from this experience that Kitahara developed his own idea that a simple mathematical theory, while beautiful, could likely be a trap. This way of thinking has proven to be a critical influence on the way he conducts his current research on biological pedigree.
The question of where life originates and how living things relate to each other has fascinated scientists and philosophers for millennia. Charles Darwin popularized the idea of a biological “Tree of Life” in his famous On the Origin of Species. Since then, scientists have updated and expanded on this idea, adding new branches and subbranches of classifications as new biological discoveries were made.
According to Kitahara, one of the most widely accepted modern incarnations of the Tree was presented by microbiologist Carl Woese in 1977. On top of introducing a new type of organism (“archaebacteria”) to the Tree, Woese developed a system of classification based on the 16S Ribosomal RNA (16S rRNA) gene sequence of an organism. 16S rRNA is a component of a cell’s ribosome, an essential part of the cell needed for protein synthesis. Ribosomes and 16S rRNA are found in the cells of all living things, and over time have diverged between different organisms due to evolutional mutations. Therefore, by mapping the 16S rRNA sequence of living organisms, scientists could count the number of discrepancies found between each organism, and by using simple math could place them on this new Tree.
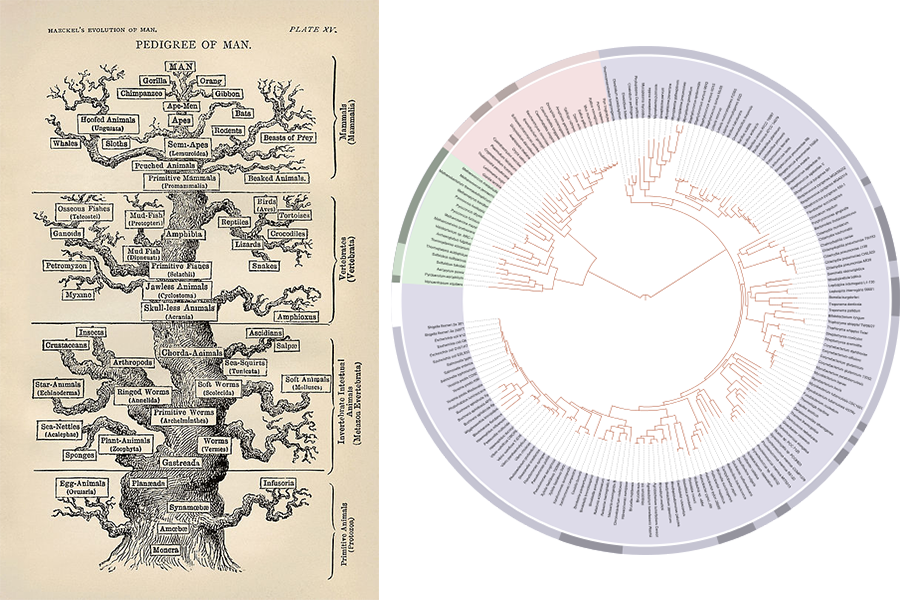
Visual evolution of the Tree of Life: German scientist Ernst Haeckel’s version as seen in The Evolution of Man (1879, left), and David Hillis’ genome-based version (2008, right). Both images taken from the public domain.
It was this mathematical simplicity in the classification of organisms that caught Kitahara’s attention.
“Woese’s idea in general is not bad,” Kiatahara says. “If a mathematician was asked to create a pedigree for organisms, they would likely use his method, as it is mathematically correct. But as I am not a mathematician, and just as I learned from my experience from my teacher’s math problem, a simple solution had to be a trick.”
In order to test whether or not Woese’s Tree of Life held up in practice, Kitahara along with his collaborator, Dr. Kentaro Miyazaki of the National Institute of Advanced Industrial Science and Technology, devised an experiment where they would completely remove the 16S rRNA sequence in an E. coli sample. They would then take a new 16S rRNA gene directly from organisms found in various sources like seawater, soils, or even fermented food, insert it directly into the E. coli sample, and observe what changes occurred in the cell’s ability to function.
“According to Woese’s premise, each organism has a 16S rRNA sequence that is unique enough to classify them as different organisms, so it therefore should be impossible to introduce a non-E. coli sequence into E. coli. However, we found that with only slight modification not only could we easily introduce a new sequence into the E. coli sample, but the new sequence could easily support the entire life activity of its new host cell. This means that because16S rRNA is essentially exchangeable between organisms, it is incorrect to strictly classify life as a tree based solely on the differences in an organism’s 16S rRNA sequence.”
The revelation that cells could survive with foreign 16S rRNA genes itself was not unknown in the scientific world. Through a process known as “horizontal gene transfer,” certain types of bacteria have been able to naturally implant their own cell’s 16S rRNA sequence into other bacteria, a process which has recently led to the development of antibiotic-resistant bacteria strains. However, Kitahara and Dr. Miyazaki have shown that this process is much more applicable to other forms of life than was originally thought.
When asked what these findings mean to the study of biological pedigree overall, Kitahara stated “As we now know that this type of transfer is possible between many organisms, I propose that rather than imagining biological pedigree as a tree, we should instead start to think of it as an inter-connected network.”
Researcher details:
Assistant Professor Kei Kitahara
Department of Chemistry
Faculty of Science
keikitahara[at]sci.hokudai.ac.jp
Author: Tyler Tannert


