Reading out RNA structures in real time
A new microscopic technique allows for the real-time study of RNA G-quadruplexes in living cells, with implications for the fight against amyotrophic lateral sclerosis.
Amyotrophic lateral sclerosis (ALS), commonly known as Lou Gehrig’s disease and Stephen Hawking’s disease, is a neurodegenerative disease that results in the gradual loss of control over the muscles in the body. It is currently incurable and the cause of the disease is unknown in over 90% of all cases — although both genetic and environmental factors are believed to be involved.
The research groups of Dr. Akira Kitamura at the Faculty of Advanced Life Science, Hokkaido University, and Prof. Jerker Widengren at the KTH Royal Institute of Technology, Sweden, have developed a novel technique that is able to detect a characteristic structure of RNA in real time in live cells. The technique, which is based on fluorescence-microscopic spectroscopy, was published in the journal Nucleic Acids Research.
“One of the genetic factors that is believed to be involved in the development of ALS is a specific sequence of RNA that forms a four-stranded structure, called a G-quadruplex,” explains Kitamura, first author of the study. “Normally, these structures regulate the expression of genes. However, a mutation in chromosome 9 in humans results in the formation of G-quadruplexes that may play a role in neurodegenerative diseases including ALS.”
One of the biggest hurdles to understanding the exact role of G-quadruplexes in disease has been the limitations in studying their formation and location within living cells in real time. The Kitamura and Widengren groups succeeded in developing a simple, robust and widely applicable technique that resolves existing issues.
The technique tracks a cyanine dye called Alexa Fluor 647 (AF647). When labeled to RNA, the fluorescence blinking state of the dye is altered with formation of the RNA G-quadruplexes. The groups analyzed the AF647-labeled RNA using a microscopy technique called TRAST (TRAnsient STate) monitoring to detect this fluorescence blinking in real time.
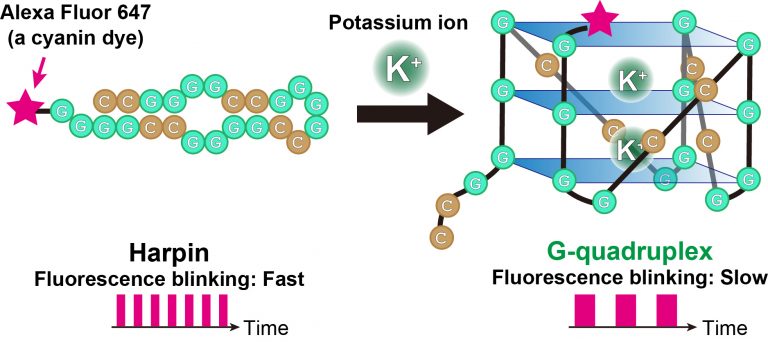
“Visually, the time-resolved changes in intensity of fluorescence appear as blinking”, said Kitamura, describing the technique. “In TRAST, we expose cells to a specific pattern of changing light intensities and measure the average intensity of fluorescence emitted from the RNA-bound dye in the cells across specific time intervals. By measuring changes in blinking properties, we can distinguish the structures of RNA within the cell.”
The team calibrated their experiment under lab conditions, determining exactly what fluorescence blinking corresponded to RNA G-quadruplexes. From this data, they were able to determine the location of RNA G-quadruplexes within living cells using TRAST.
This work proves that cyanine dyes can provide sensitive readout parameters on the folding states of RNA G-quadruplexes in living cells, and even for single cells. This, in turn, allows for the possibility of studying the RNA G-quadruplexes in disease in real time at intra-cellular level. It can also be applied to study the folding and misfolding of proteins in cells.

Original Article:
Akira Kitamura, Johan Tornmalm, et al. Trans-cis isomerization kinetics of cyanine dyes reports on the folding states of exogeneous RNA G-quadruplexes in live cells. Nucleic Acids Research. January 18, 2023.
DOI: 10.1093/nar/gkac1255
Funding:
This research was supported by the Swedish Research Council (VR 2017-04057, VR OQS 2016-06122); the Knut and Alice Wallenberg Foundation (KAW); the Swedish Foundation for Strategic Research (SSF, ITM17-0491, BENVAC RMX18-0041); a Japan Society of Promotion of Science (JSPS) Grant-in-Aid for the Promotion of Joint International Research “Fostering Joint International Research” (16KK0156), a Grant-in-Aid for Scientific Research (18K06201, 22H04826), Grants-in-Aid for Scientific Research on Innovative Areas “Chemistry for Multimolecular Crowding Biosystems” (JP20H04686) and “Information physics of living matters” (JP20H05522); the Japan Agency for Medical Research and Development (AMED, JP22gm6410028, JP22ym0126814); the Canon Foundation Research Grant Program “Creation of Industrial Infrastructure”; the Hoansha Foundation in Japan; and the Hokkaido University Office for Developing Future Research Leaders (L-Station).
Contacts:
Dr. Akira Kitamura
Laboratory of Molecular Cell Dynamics
Faculty of Advanced Life Science
Hokkaido University
Tel: +81-11-706-9542
Email: akita[at]sci.hokudai.ac.jp
Sohail Keegan Pinto (International Public Relations Specialist)
Public Relations Division
Hokkaido University
Tel: +81-11-706-2186
Email: en-press[at]general.hokudai.ac.jp
